This is an interesting article that I actually stumbled across a citation to while reading a book (Genetics and the Logic of Evolution, Kenneth Weiss and Anne Buchanan). It is several years old now.
"Functional proteins from a random-sequence library." Keefe, A.; Szostak, J. Nature, 2001, 410, 715-718.
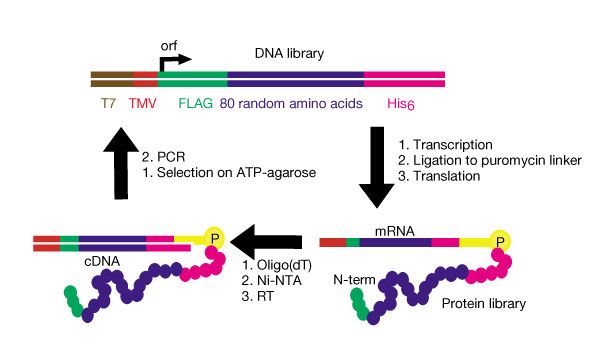
The authors were interested in how frequently a random library of peptides would include a useful protein.
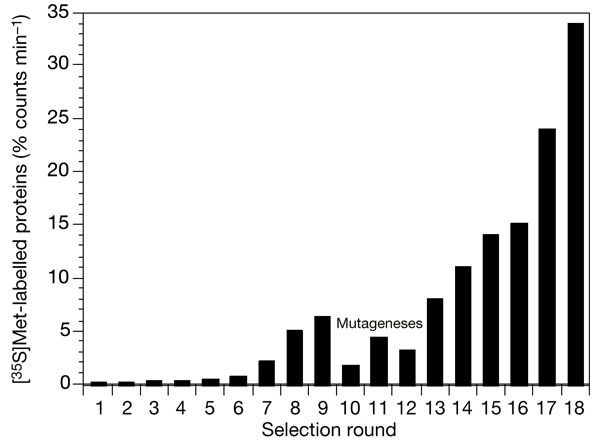
After eight rounds of this they had 6.2% of the proteins binding ATP. These were found to belong to four different families, each originating from one ancestor protein present in the random pool, and had no resemblence to existing ATP-binding proteins or other proteins. These were then subjected to error-prone PCR to mutate the proteins further. After six more rounds of mutagenesis the ATP-binding fraction rose to 34%. In this process three of the four original families were wiped out, and all proteins present belonged to one of the original families.

"Functional proteins from a random-sequence library." Keefe, A.; Szostak, J. Nature, 2001, 410, 715-718.
The authors were interested in how frequently a random library of peptides would include a useful protein.
In order to test this they made a humongous library of peptides linked to their mRNAs. They converted the mRNAs to DNAs, ran the DNA/protein complex through ATP-agarose to find those that bound ATP, amplified these by PCR, and repeated the process.Functional primordial proteins presumably originated from random sequences, but it is not known how frequently functional, or even folded, proteins occur in collections of random sequences. Here we have used in vitro selection of messenger RNA displayed proteins, in which each protein is covalently linked through its carboxy terminus to the 3' end of its encoding mRNA (1), to sample a large number of distinct random sequences. Starting from a library of 6 x 10^12 proteins each containing 80 contiguous random amino acids, we selected functional proteins by enriching for those that bind to ATP. This selection yielded four new ATP-binding proteins that appear to be unrelated to each other or to anything found in the current databases of biological proteins. The frequency of occurrence of functional proteins in random-sequence libraries appears to be similar to that observed for equivalent RNA libraries (2, 3).

After eight rounds of this they had 6.2% of the proteins binding ATP. These were found to belong to four different families, each originating from one ancestor protein present in the random pool, and had no resemblence to existing ATP-binding proteins or other proteins. These were then subjected to error-prone PCR to mutate the proteins further. After six more rounds of mutagenesis the ATP-binding fraction rose to 34%. In this process three of the four original families were wiped out, and all proteins present belonged to one of the original families.

Comparing the round 18 sequences with the ancestral sequence showed that four amino-acid substitutions had become predominant in the selected population (present more than 39 times in 56 sequences, Fig. 3b), and that 16 other substitutions had also been selectively enriched (present more than 4 times in 56 sequences, Fig. 3b). In addition, each clone contained a variable number of other substitutions. The selectively enriched substitutions are distributed over the 62 amino-terminal amino acids of the original 80-amino-acid random region, suggesting that amino acids throughout this region are contributing to the formation of a folded structure, at least in the complex with ATP. The substitutions in each of the assayed clones improve ATP-binding relative to the ancestral sequence.
This is interesting--none of the other proteins had this zinc-binding motif. They did not add zinc to the selection buffer, but said that it was present in low levels in mammalian blood which they used to prepare the lysate to translate the mRNAs. They think this indicates that metal chelation provides an easy way to induce folding in simple proteins.These (family B) proteins contain two invariant CXXC sequences (56/56 clones sequenced) within the core domain. Also, protein 18-19 is functional in the presence of 5 mM dithiothreitol, indicating that disulphide bonds are probably not required for ATP binding. In addition, a deletion construct removing only one of these cysteines did not bind ATP (Fig. 3c). These observations suggested that family B proteins might require a coordinated metal ion to be functional. Elemental analysis by atomic absorption spectroscopy revealed the presence of one equivalent of bound zinc, and no other divalent metals. Incubation of the protein with EDTA results in a concentration-dependent loss of ATP-binding activity. Activity can be restored to protein that has been extensively dialysed in the presence of EDTA by the addition of Zn^2+, but not Mg^2+, ions.
Thank goodness that's not me.In conclusion, we suggest that functional proteins are sufficiently common in protein sequence space (roughly 1 in 10^11) that they may be discovered by entirely stochastic means, such as presumably operated when proteins were first used by living organisms. However, this frequency is still low enough to emphasize the magnitude of the problem faced by those attempting de novo protein design.
